Projects
The HackTrack is the hacking component of a Brainhack event, where people can work together on projects. For tutorials and learning sessions check the TrainTrack page.
What projects? Any kind! From exploding brains to resource gathering and data sharing!
i spy with my little eye
The goal of this project is to adapt or develop a process that helps communities create a controlled vocabulary for assessment tools that are used in the community.
Short definitions here:
- an assessment
- is a questionnaire, named form, cognitive or clinical assessment or battery of test, observational protocol
- is not a cognitive task (n-back), experimental design or condition (medication on/off), biological or physical measure (height, blood pressure)
- a controlled vocabulary is a list of terms with definitions and unique identifiers (e.g. the ICD-11)
- a community is a group of people with data that agree to work together and could benefit from a shared language for their assessments (e.g. a research consortium, a data publishing platform, a group of collaborators, ...)
Assumptions:
- Most communities collect “assessment tools”, questionnaires, forms, clinical assessments, etc
- Within a community, agreeing on a well defined list of named assessments is good for interoperability, findability, etc
- There are many controlled FAIR vocabularies that include assessment tools
- Making a single controlled vocabulary of assessments that works for all communities is not feasible
- A community vocabulary of assessments should link to existing FAIR vocabularies so we may eventually integrate communities etc
- There are good tools / services out there that can help find terms in existing FAIR vocabularies (OLS, UMLS, ...) and we may reuse these
Goals
Adapt / create (a) process(es) that
- lets a community collect, curate, and maintain a list of assessments
- makes it easy for community members to propose new terms
- facilitates / requires new terms to link to terms in existing FAIR vocabularies
- makes the review of new submissions easy / manageable
- helps publish and maintain these lists in a format that is easily reusable
The link to Neurobagel here is very loose, but we help communities annotate their data with FAIR vocabularies - and the insufficient coverage of assessment tools is the main complaint we are getting. Eventually the goal would be to allow communities to create their lists and for Neurobagel to be able to read these lists for annotation and discovery.
Nipoppy pipeline catalog
Nipoppy is a neuroinformatics framework for standardized organization and reproducible processing of neuroimaging-clinical pipelines. We have recently launched a community-driven pipeline catalog on the Zenodo data repository. Users can use the Nipoppy command-line interface to search the pipeline catalog, download available pipelines and upload new pipelines. We propose two projects related to the pipeline catalog, as well as an additional project for more advanced brainhackers.
Subproject 1: Improve Zenodo interface Want to contribute to an open-source software project? Help improve the Nipoppy CLI’s Zenodo interface! We have identified a list of good first issues for newcomers.
Subproject 2: Contribute new pipelines to the pipeline catalog Have a pipeline you would like to share? Want to use Nipoppy to run a pipeline that is not currently in the pipeline catalog? Help extend the pipeline catalog by adding new pipeline(s) to it! This project does not require any Python programming skills.
Subproject 3: Add telemetry to the Nipoppy CLI Telemetry allows for the recording of usage statistics for software tools, which can be useful to demonstrate the usage level/impact of a tool. Join this project to help design and implement telemetry to the Nipoppy CLI.
From Nipype to Pydra: Modernising Workflow Engines for Neuroimaging
The neuroimaging community has long relied on Nipype to construct reproducible, modular workflows (e.g. fMRIPrep, MRIQC, dMRIPrep). While Nipype has been foundational, it carries significant technical debt: a complex execution model, outdated dependencies, and no support for dynamic task graphs. Now, with the alpha release of Pydra—a modern, Pythonic, async-ready successor first conceived at OHBM Brainhack 2018—we have the opportunity to transition to an easy to a developer-friendly, maintainable and powerful workflow engine.
This Brainhack project aims to help the field get over the activation energy needed to make Pydra the obvious choice for neuroimaging workflows, and move closer to formally sunsetting Nipype. To do this, we’ll focus on closing key feature gaps (e.g. a MATLAB Compose module, a command-line interface), migrating core interfaces and workflows, and improving documentation to support new users.
Whether you're a long-time Nipype user, an interface developer, or entirely new to workflow engines, there’s a meaningful way to contribute.
-
Matlab Compose Module for Pydra Add a
pydra.compose.matlabmodule to allow integration of MATLAB tasks, similar to Nipype’s MatlabCommand. This will allow seamless inclusion of SPM or other MATLAB-based tools in Pydra workflows. -
Command-Line Interface for Pydra Design and prototype a simple CLI (
pydra run ...,pydra crash ...) for defining and executing workflows from the terminal. This may use click, typer, or similar libraries, and could support YAML/JSON-based workflow definitions. -
Workflow Visualization Integration Inteface Pydra with the PIM-V3 visualizer workflow visualiser tool.
-
Nipype → Pydra Migration Tools Use the nipype2pydra converter to partially migrate Nipype interfaces and workflows into their Pydra equivalents, then test and manually edited the results.
-
Fresh Eyes on Documentation As a newcomer to Pydra or to workflow engines, your perspective is invaluable! Help improve onboarding materials, identify confusing sections, and contribute to or annotate the official Pydra docs.
EasyBCI: Cross-platform General Purpose EEG Interface for Game and Application Inputs
- Python
- In-person (Brisbane)
- Machine-learning
- EEG
- BCI
This project develops a customization interface that translates EEG signals into computer inputs. Users can train the system to recognise distinct brainwave patterns (mental/motor imagery), each of which corresponds to a configurable metric/class. These metrics are then dynamically mapped to specific computer keys (or mouse input), enabling EEG-based interaction with games, applications, or custom software environments.
Behind the scenes is the EEGNet classifier. Find out more about the classifier here. The aim is that this application will train the model locally on the users machine, and use it to predict the class of the EEG data, and then map it to the a computer input (keyboard or mouse). The user can then theoretically use their EEG to control any program on their computer, and have full control over the interface. We currently support the Neurosity Crown and the OpenBCI Cyton devices, but this can be easily expanded to more. This project is committed to free and open source practices for it's entire lifecycle.
Nilearn: Statistics and Machine Learning for Neuroimaging in Python
Nilearn is an open-source Python package for fast and easy analysis and visualization of MRI brain images. It provides statistical and machine-learning tools, with instructive documentation and a friendly community. It includes applications such as multi-voxel pattern analysis (MVPA), decoding, predictive modelling, functional connectivity, and brain parcellations.
Recently, we did some analysis of the package's usage (https://github.com/nilearn/poia) and found out that "nilearn.plotting" is the most used module. Therefore, we want to dedicate this year's Brainhack to improving the plotting module by resolving existing issues, welcoming new ones, and improving the documentation. To this end, we encourage users to simply filter the issues by the "Plotting" label (https://github.com/nilearn/nilearn/labels/Plotting). In addition, if you are motivated to work on any other issues, we welcome that as well! We have a lot of open issues in the repository, and we would love to see contributions from the community. New contributors should look for the "Good First Issue" label to get started (https://github.com/nilearn/nilearn/labels/Good%20first%20issue).
Bayesian modeling of structural connectivity
- Python
- R
- In-person (Brisbane)
- Bayesian
- git
- Probabilistic Programming
- Diffusion MRI
- Structural Connectivity
The goal with this project is to develop an atlas of the reliability of structural connectivity matrices based on Bayesian modeling. That model should fulfill the following two objectives: 1) quantify the reliability of fiber density estimation for each connection, 2) obtain a probability indicating whether the connection is most likely absent or present. I have developed a first model implemented in the Python library PyMC, but this model needs improvement.
Goals:
- Modify the current model to generate a good estimate of the probability that a connection is truly absent or present, based on simulated repeated measurements of a structural connectivity matrix. Possible leads include regularization, modify parameter prior distribution and modify model implementation.
- Incorporate connection length in the model
- Fit that model on real structural connectivity matrices and interpret the results
See this abstract for more details about the current status of the project: https://www.overleaf.com/read/zhsnjpskpzzh#d2befe
NeuroViz Graph Gallery
- Python
- In-person (Brisbane)
- Tutorials/Examples
- Vizualization
We aim to build a community‑driven hub of scriptable neuroimaging visualizations (initially in Python). It will feature example scripts organized by visualization type (via a simple taxonomy), so you can quickly find exactly what you need. Think of it as a Python Graph Gallery for brain data: intuitive, code‑ready, and powered by the community. Down the road, we hope to welcome contributions in other popular languages like R and MATLAB, too.
Have a favorite tool or snippet? Pull requests and suggestions are more than welcome!
PsyR - an R package for better inference in multivariate statistical analysis
- R
- In-person (Brisbane)
- documentation
- Numerical analysis
- Multivariate statistics
- Pascal
We consistently use massive datasets across neuroscience and psychology. The routine gathering of big data requires that we are well equipped with tools that allow us to conduct appropriate multivariate statistics. This project aims to produce an R package that allows the researcher to overcome little discussed limitations of traditional multivariate statistical analyses.
Multivariate statistical analysis (e.g. MANOVA and repeated-measures ANOVA) typically follows a two stage procedure; an omnibus test of the global null hypothesis followed by post-hoc tests of specific effects. It is not well known that under certain circumstances, such as when the omnibus test is overpowered, that the type 1 error rate for this procedure is drastically inflated, sometimes to a type 1 error rate of 1! It is even less well known that this procedure can lead to an even lessor known type IV error, which is the incorrect interpretation of a correctly rejected hypothesis. This is caused when the follow-up contrasts are inadequate to test the question of interest, as can occur when testing simple effects.
It is possible to avoid these dragons by using an alternative procedure where all inferences are derived from simultaneous confidence intervals (SCIs) on contrasts of interests. The 'simultaneous' bit means that the same statistic contributes to both the omnibus and the contrast tests for significance, which controls the type 1 error rate. Even better, computing confidence intervals on contrasts of interests allows reseachers to move away from binary decision-making (is something significant or not?) to interpretations involving magnitude (how big is this effect likely to be at the population level?).
One piece of software (PSY) can produce SCIs appropriate for both planned analyses (where contrasts are defined independently of the data) and for more flexible analyses where contrasts are defined on a post-hoc basis. However, this software is only available for use on windows and cannot be scripted into reproducible workflows. We have, over several Brainhacks, built an R package that implements the functions of PSY, and to make this method of statistical inference available to the masses!
Our goals for the OHBM Brainhack 2025 are:
- Unlock the final fancy function of Psy; converting from pascal code to an R function the approximation required to compute the cdf of a particular distribution needed to calculate test statistics for 2 x 2 balanced ANOVA designs.
- Write vignettes and help documentation for the package's functions so far.
- Explore packages and ways to make contrast definitions easier for novel users.
- Add Scheffe contrast functionality to the R package.
- Add/ensure contrast scaling functions for appropriate interpretations of output.
Learning to create GUIs on tkinter
- Python
- Hybrid (Asia/Pacific)
- In-person (Brisbane)
- GUI
- tkinter
A while back I tried to create a Graphical User Interface for a set of python toolboxes of the Physiopy Community. After looking into different options, I ended up playing with tkinter to approach the creation of GUIs, and started putting together a few elements to interact with the main toolboxes. The first very draft-y result of this play session was this, and while someone might find it pretty (ugly), for sure it's not functional, as I did not associate yet functions to the GUI elements itself. It takes time to learn a new skill!
So I thought: what best place to learn new hacking skills than the Brainhack! If you are interested in learning some GUI programming in tkinter with me, please join me! We can develop GUIs for Physiopy's toolboxes together, or if you have your own toolbox we can just learn and compare our work and help each others to get out of this Brianhack with a semi-functional GUI toolbox!
QCApp - Last mile development
- Python
- R
- In-person (Brisbane)
- git
- Data Science
- Quarto
QCApp is a new application for conducting manual QC of MRI preprocessing. It is heavily inspired by previously successful frameworks such as brain_match, braindr, QRATER, and also any pipeline that generates html reports (such as fmriprep). QCApp aims to be an extensible platform for quickly reviewing several different kinds of neuroimaging pre-processing steps locally. The basic workflow facilitated by the app is simple: view informative, visual displays that allow determination of the success of neuroimaging processing steps, structure the displays so that people can rate thousands of images quickly, and provide an efficient system for recording and reviewing those ratings.
Here is a slidedeck pitching QCApp as a whole: pitch.pdf. The deck includes some long-term goals for QCApp, but the hackathon will focus on projects with a narrower scope.
Currently a basic draft of QCApp is available. With lots of hacking, it could be run on a small dataset. But no one has time for that! Fortunately, use of the app could be greatly simplified if either of the following sub-projects were completed:
Develop helper CLI for creating rating database
- QCApp displays visual summaries of preprocessing steps (e.g., a transparent overlay of a mask on the to-be-masked anatomical image, which allow determination of whether the mask is too conservative or too liberal). But it does not itself generate these visual summaries. This project involves creating a command line utility that can generate the images to rate.
- The linked issue includes several gists that could form the basis of this utility.
Create basic dashboard for viewing ratings
- After rating several images, it should be simple to identify which steps might have failed. This project involves creating a simple dashboard for determining preprocessing quality.
- Ratings are stored in an sqlite database (along with the images that were rated), and so it is expected that the dashboard will be based on the tables within that database.
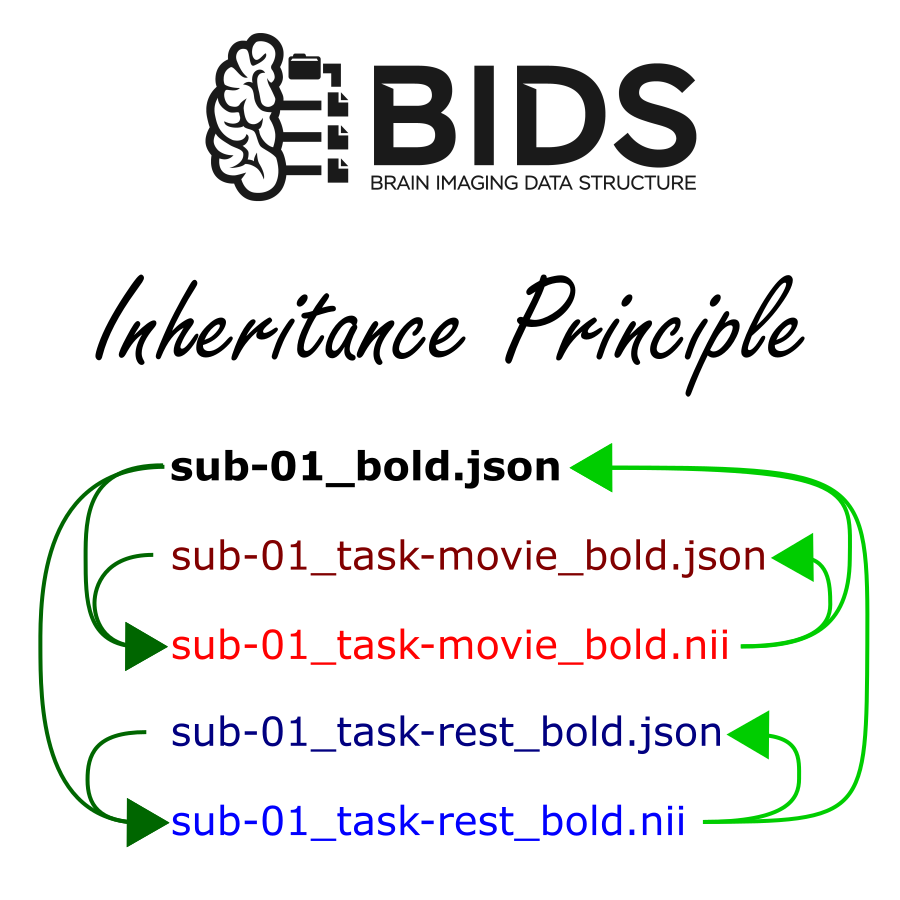
IP-Freely: Free manipulation of BIDS datasets according to the Inheritance Principle
- In-person (Brisbane)
- BIDS
- BIDS App
- git
The Inheritance Principle (“IP”) has been a feature of the Brain Imaging Data Structure (BIDS) since its inception. It describes how the metadata relevant to one specific data file may in fact originate from multiple metadata files, and how metadata defined in one location may apply to multiple data files. These can both reduce redundancy and communicate the intrinsic hierarchical structure of the dataset. This however comes at considerable complexity to the specification itself and the software responsible for interfacing with such data. Indeed nearly a decade after its creation, many software APIs for parsing BIDS data still do not support the Inheritance Principle.
There is a dearth of existing software tools for managing the IP. If a dataset that exploits the IP were provided as input to a BIDS App that is naive to the IP, that App could perform improper association of metadata. Existing datasets may contain substantial metadata redundancy, due to not currently having an easy way through which to exploit the IP.
The future of the IP is at a crossroads, particularly as work progresses toward BIDS 2.0:
- On one hand, some would like for the IP to disappear entirely: all metadata relevant to any given data file would need to be stored in a sidecar metadata file alongside that data file, regardless of the extent of duplication.
- On the other, not just preserving the IP but augmenting it could better communicate the complex hierarchical dependencies of complex datasets, particularly derivative data.
- The potential impact that the existence of the Inheritance Principle has (or could have) had on existing datasets is impossible to ascertain.
The goals of this project are:
- Commence construction of a BIDS App that is responsible for:
- Detecting the presence or absence of the Inheritance Principle in any given BIDS dataset
- Manipulating that dataset to either remove all uses of the Inheritance Principle or make maximal use of the Inheritance Principle under some ruleset.
- Manually create a dataset that encapsulates all of the different manifestations of the Inheritance Principle, with multiple instantiations with and without use of the Principle. This will then act as a reference dataset against which the operation of the software tool can be verified.
Multi-echo fMRI data analysis Jupyter book
- Python
- In-person (Brisbane)
- Jupyter
- fMRI
Multi-echo fMRI is increasingly popular and is used in at least 40 presentations and posters at OHBM2025. Optimizing acquisition parameters and planning data analyses benefits from a high-level understanding of MRI physics and processing methods. Existing centralized resources for this training are currently limited. Several years ago, @tsalo started a Jupyter Book on "Multi-echo fMRI data analysis" , but there are both known gaps and opportunities to contribute new ideas.
A good example page of resources that can be created in a Jupyter book is here: https://me-ica.github.io/multi-echo-data-analysis/content/Signal_Decay.html
Goals
- Add content with a focus on improving the sections on Theoretical background, Practical Resources, and Analysis tutorials.
- Copy/move general educational material from https://tedana.readthedocs.io/en/stable/multi-echo.html software documentation into the book.
Benefits from contributing
- Learn more about multi-echo fMRi and multi-echo fMRI methods
- Learn how to publish via a Jupyter book & ways to share useful visualizations within this format (Full disclosure: The project lead hasn't written a Jupyter book before & is also hoping to use this as a motivation to gain this skill)
Note 1: This could potentially overlap with #10 and multi-echo fMRI neuroimaging examples designed for that project could be part of this book.
Note 2: Whether or not this project happens will depend on who else is at the brainhack and their general interests. Other ideas I considered submitting and would consider helping lead are:
- Working on integrating NiiVue into tedana's Interactive Reports with the goal of gaining a better understanding how to add NiiVue to a range of packages
- Working on visualizing and summarizing better data quality metrics for fMRI
An Interactive Neuroimaging Learning Platform
- Bash
- Python
- In-person (Brisbane)
- NeuroDesk
- Jupyter
Training in neuroimaging faces a persistent challenge: existing tutorials often lack interactivity and quickly become outdated as software evolves, leaving researchers with broken workflows and steep learning curves. This project addresses these issues by developing an open repository of interactive, automatically tested Jupyter notebooks that demonstrate neuroimaging workflows using Neurodesk - a portable, container-based neuroimaging platform. This would allow users to explore parameters hands-on while building foundations for their own analyses.
Goals
- Develop & Test interactive neuroimaging examples (e.g. from FSL course or Andy's Brain Book)
- Platform: Jupyter Notebooks, GitHub Actions, and Neurodesk
Community Impact: This framework has potential for significant growth through contributions from global Brainhack events, enriching the resource pool with diverse analysis techniques and supporting a broad range of neuroimaging training needs.
Neurodesk container builder
- In-person (Brisbane)
- documentation
- NeuroDesk
- Containers
Our team has made it easier than ever to share your tool with the wider community. With an interactive browser interface, we allow users to create a setup instruction for a tool that will be containerized by our builder and included in the Neurodesk environment. The project aims to:
- Test the usability of the builder interface
- Enhance documentation from the users' perspective
- Share cool tools with others through Neurodesk
Repronim Inventory
- Any programming language
- In-person (Brisbane)
- Webdev
- Tutorials/Examples
Over the years, multiple cohorts of ReproNim Fellows have developed a diverse collection of training materials and educational resources (“training products”). We aim to organize, curate, and make these resources publicly accessible for broader community benefit.
Goals for the Brainhack project: • Goal 1: Complete the sorting and categorization of the ReproNim training products to create a structured, searchable collection (at the moment in a google sheet) . • Goal 2: Brainstorm and prototype a platform or website that enables users to browse, filter, and explore these resources based on tags such as topic, format, or audience.
Network Level Analysis of Connectome-Wide Associations
- Matlab
- Hybrid (Americas)
- In-person (Brisbane)
- Network analysis
This is a newly released toolbox for performing brain connectivity-behavior analysis with statistical inference at the network-level. The current toolbox includes canonical area/network parcellations in adult and pediatric cohorts and offers a variety of statistical testing options supporting single group or between group comparisons. Our goal is to get feedback from potential users to maximize the toolbox's usability among the neuroimaging community. Goal 1: Get feedback on usability for most basic use case (use GUI on provided data to run model and view results) Goal 2: Run GUI software with custom connectivity and behavior/clinical outcome data from user
Neurosynth Compose: neuroimaging meta-analyses for a(llm)
- Python
- In-person (Brisbane)
- documentation
- meta-analysis
Our team is making neuroimaging meta-analyses quicker and easier. We have a platform for you to search and curate studies, specify meta-analyses, and execute/record your results. This year we used large language models to extract information from studies and display the results to users. The projects/goals this year are:
- Test the new features we have added
- Suggest/implement pipelines for extracting information from papers
- Add documentation for the extraction pipelines
- Give feedback on tutorials
- extract coordinates from pdf files in semantic scholar