OHBM Brainhack
June 16-18th 2021
What is it Register Submit your project Sign up for hands-on sessionsAbout OHBM BrainHack 2021
The event will take place on Sparkle! Every registered participants will receive the credentials via email, if you did not get the email please get in touch!
What is OHBM? OHBM is the Organization for Human Brain Mapping. A Society of scientists dedicated to mapping the brain using all neuroimaging technologies.
What is Brainhack? An event where people get together to learn about open science and to collaborate on projects they’re interested in - it can be on anything!
The OHBM Brainhack is a chance for anyone interested in brains to work collaboratively on common projects (during the hacktrack) and learn about new ideas and tools in open science (during the traintrack).
Brainhacks are not the typical academic conference. Here, attendees can actively take part in the program and learn from each other - sounds something you would like? You can become a part of the event as a volunteer filling this form. Many Brainhack projects might involve coding, but it is not a requirement and you can contribute in most cases even without coding skills.
We look for neuroimagers of all modalities from all over the world at different career stages, and coming from all racial and gender backgrounds who are interested in working together with others to build an open science community. If you're new to the Brainhack community, please join us! If you belong to any minority groups we want to reiterate that we want you here and are excited to have you join us!
Share Ideas

Share your research and curiosity with other neuroimaging and computer science researchers.
Build Projects

Try the presented tools on provided data or bring your own tools/data!
Be social
Get to meet and socialize with people from all over the world!
Learn Tools

Learn about brain imaging data and how to apply the latest computational tools to your data.
What's Up? #OHBMHackathon
HackTrack projects
The HackTrack is the official fun side of a Brainhack event, where people can work together on projects. What projects? Any kind! From exploding brains to resource gathering and data sharing!
Would you like to propose a project? Just open an issue on our GitHub repository and fill the template, we will be in touch to help you get going! But be sure to register first!
Do you plan to focus on visualization? Are you getting images so weird that they are kind of beautiful? Consider participating in the output-of-hackathon competition organized by the BrainArt SIG! You can find the submission form here.
And one more thing: as this year we are sponsored by QMENTA, you can take advantage of their platform to run your project using resources in the cloud! Do you want to know more about it? You can check out this video or you can get in touch directly with them on their Brain Innovation Hub Slack!

NeuroDesk - A scalable and easy to use data analysis environment for reproducible neuroimaging
Time slot : Rising sun
PROJECT DESCRIPTION
Neuroimaging researchers require a diverse collection of bespoke command-line and graphical tools to analyse data and answer research questions. Installing and maintaining a neuroimaging software setup is challenging and often results in un-reproducible environments. Container technology, such as docker or singularity, enables the execution of software on different operating systems and could aid in distributing scientific software. We are developing NeuroDesk, a platform build on container technology for processing and analysing neuroimaging data with the aim to lower the barrier of using various neuroimaging software in a reproducible environment, so that researchers spend less time on setting up and maintaining an analysis environment.
More information in this github issue
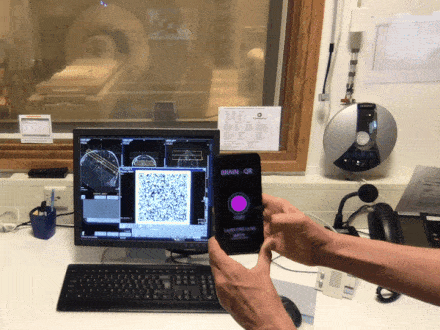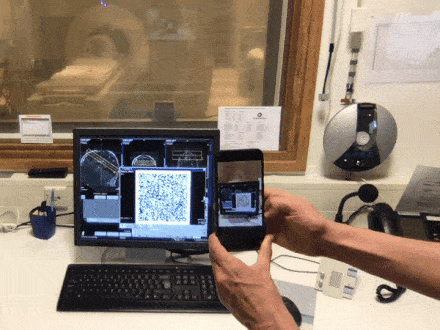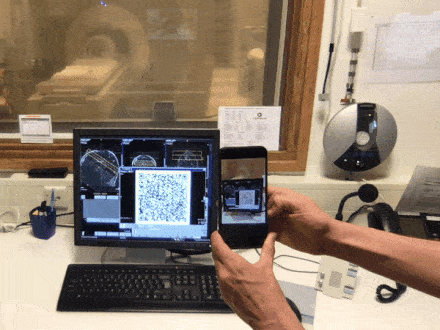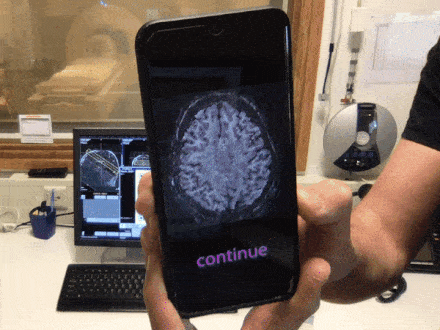
Brain QR Modem - A setup to stream MRI images unidirectionally to the cloud via QR code animations
Time slot : Atlantis
PROJECT DESCRIPTION
Have you ever been annoyed how hard it is to get brain data off the scanner? The fact that scanners usually contain private information about patients and are thus embedded in maximally restrictive clinical cyber-security environments, makes it quite complicated to get access to the data. Especially when visiting collaborative sites. In this Hackathon project, we aim to develop a purely uni-directional (safe) data streaming “hack” to transfer MRI data directly to the cloud by means dynamic QR codes. In the early days of the Internet, modems (modulator-demodulator) were used to (i) convert digital information into audio streams, (ii) transfer them across telephone lines, and (iii) convert them back into the digital domain. Here, we aim to do the same thing with pixel data of MRI scans. However, instead of audio signal we will use machine-readable visual information: QR codes.
More information in this github issue

Flooding brains: Mesmerizing & relaxing volumetric brain animations
Time slot : Atlantis
PROJECT DESCRIPTION
After 🎆 exploding brains 🎆 in the last year's brainhack, now it is time for animations of a more relaxing sort. I will be mainly using a few LayNii programs that I have been developing for high resolution imaging over the last year (e.g. layers, columns) to generate mesmerizing brain animations. I will focus on using animal brains from one of my favorite projects: https://braincatalogue.org/.
More information in this github issue

BIDS Execution Specification
Time slot : Atlantis
PROJECT DESCRIPTION
An extension proposal that has been in the works for a while, and builds upon the original BIDS Apps paper, this extension seeks to further standardize the execution specification for BIDS applications, including integration with Boutiques.
More information in this github issue

Phys2BIDScoin: Integrating BIDScoin and phys2bids for a user-friendly GUI based BIDS conversion of physiological files
Time slot : Atlantis
PROJECT DESCRIPTION
Phys2bids is a python3 toolkit meant to format physiological files in BIDS. BIDScoin is also a python3 toolkit that is meant to format MRI source files in BIDS. BIDScoin comes with a data discovery routine and a graphical interface to customize the formatting heuristics. The actual data discovery and formatting is done using plugins and the BIDScoin framework itself is agnostic about the source modality. Phys2bids currently lacks data discovery functionality and BIDScoin lacks a formatting backend for physiological data. The goal of this project is to develop the plugin interface between BIDScoin and phys2bids and hence combine their complementary functionality for both projects.
More information in this github issue
Physiological signal classification challenge
Time slot : Atlantis
PROJECT DESCRIPTION
Phys2bids is a python3 library to format physiological files in BIDS. One of the current development goals is to make phys2bids smarter by implementing an automatic signal classificator. Ideally, given a physiological signal as input, phys2bids should be able to determine the type of the signal (cardiac, respiratory …).
To move forward, this project is structured as a challenge: we provide time-series data of 4 kinds of physiological signals (cardiac, respiratory_chest, O2 and CO2) and the goal will be to collaborate to find robust features that allow discerning between them.
More information in this github issue
Brainhack book: all the shenanigans
Time slot : Atlantis+Rising sun
PROJECT DESCRIPTION
Brainhack Jupyterbook is an growing resource originated from the conception of the 2021 Neuroview paper. Currently, the book contains companion material of the paper, but we are looking forward to hear what other content the community want it to have. Join the discussion!
More information in this github issue

Social gather.town for the hackathon and OSR
Time slot : Atlantis+Rising sun
PROJECT DESCRIPTION
We aim to decorate the open science gather.town room for the open science activities and set up something fun for the OSR social.
More information in this github issue

ARTEM-IS web app: working towards an Agreed Reporting Template for EEG Methodology (International Standard)
Time slot : Atlantis+Rising sun
PROJECT DESCRIPTION
The dense data streams and analytical flexibility of electroencephalography (EEG), result in a substantial ‘Garden of Forking Paths’. Recent research (Šoškić et al 2021) shows that most research reports do not contain sufficient detail in their methods to know which path has been taken, hampering reproducibility, replication and meta-science. Our initiative to create an Agreed Reporting Template for an International Standard (ARTEM-IS) includes a draft template designed to make reporting EEG methodology easier and more accurate, by providing fields for researchers to input specific methodological details. Read more about why we need this tool and how you can help to make it better in our (preprint).
More information in this github issue

The courtois-neuromod physiological data as a use-case for phys2bids
Time slot : Atlantis
PROJECT DESCRIPTION
Phys2bids provides a way to convert physiological recording files to bids format. This brainhack project acts as user testing for the software by applying its workflow to a large-scale dataset. The Courtois Neuromod project is a longitudinal program which collects massive amounts of multimodal data (neuroimaging, peripheral physiologicals signal and eye-tracking) with the same 6 subjects throughout different tasks. We want to apply the phys2bids workflow to the entire physiological signals (electrocardiographic, electrodermal, respiration and pulse) dataset.
More information in this github issue
BIDS-Prov Specification
Time slot : Atlantis
PROJECT DESCRIPTION
A BIDS extension proposal to represent what pipeline was applied to a given dataset.
More information in this github issue
Developing collective action campaigns for the neuroimaging community
Time slot : Rising sun
PROJECT DESCRIPTION
Academia is trapped in a collective action problem: we all know there are better ways we could be doing things, but feel pressured to 'play the game' in order to survive a competitive career in science. Historically, collective action problems like this have been solved with collective action, i.e. the mass uptake of new behaviours that aim to further the group's interests. More recently, online conditional pledge platforms (e.g., Kickstarter) have demonstrated that collective action can be organised on a global scale with zero risk to users, serving to cut out middle-men and directly benefit the users themselves. At present, however, this strategy has yet to take hold in academia.
Project Free Our Knowledge (FOK) aims to rectify this by organising collective action in the research community. Using our platform, researchers can pledge to support open science practices, subject to there being a critical mass of support for the proposed behaviour. If the threshold is achieved, everyone who has pledged is contacted and directed to carry out the action together, thus motivating cultural change with minimal risk to individuals.
More information in this github issue

FLOW - Integrating EEG analysis scripts in a cloud-based database management system
Time slot : Atlantis
PROJECT DESCRIPTION
Brain Electrophysiology Laboratory is proud to present FLOW (Forward Looking Operations Workflow), an all-in-one solution for EEG database management, ERP analysis pipelines, and visualization with AWS cloud or on-premises server implementations. We use containerization technology to run ERP analysis Python scripts as modular units with I/O to the FLOW database, thus facilitating reproducibility across different stages of analysis. The containerization approach allows easy integration of existing analysis scripts, thus making FLOW an environment the can be adjusted to lab needs.
More information in this github issue
Automated evaluation of quality metrics for brain data using deep learning
Time slot : Atlantis
PROJECT DESCRIPTION
The goal of this project is to create an automated deep-learning based pipeline for evaluation of quality metrics for 3D brain imaging data and providing a decision on the quality and usability of the data.
More information in this github issue
Nobrainer: a python framework for neuroimaging data
Time slot : Atlantis
PROJECT DESCRIPTION
Nobrainer is a deep learning toolbox for 3D neuroimaging data analysis. It includes implementation of many deep learning tools including data preprocessing pipelines, models structures, trained models, etc. In this hackathon project we are going to develop various aspects of this toolbox. This project has different parts depending on your skills. It spans from data curation to adding new implementations to the toolbox.
More information in this github issue
Fuzzy libmath, transparently evaluating the numerical stability of your pipeline against the elementary mathematical functions (libm)
Time slot : Atlantis
PROJECT DESCRIPTION
Fuzzy libmath is evaluating the pipeline stability by applying noise to mathematical functions using the library call interposition technique. This project aims to study whether the Monte Carlo Arithmetic method is a truly good perturbation model for evaluating pipeline stability across the operating systems.
More information in this github issue
Nipype 2.0: building new ecosystem around Pydra
Time slot : Atlantis
PROJECT DESCRIPTION
We are creating Nipype 2.0 ecosystem around Pydra package. Pydra is a new dataflow engine, coordinating the movement of data from the output of one computational node to the input of another node. A workflow is represented as a graph, and each node is a Task object, which may be a Python function or an interface to an external tool.
During this hackathon we want to work on the core package, but also prepare converters for Nipype 1.0 interfaces. We are also planning to improve documentation and tutorials.
Pyrda is a lightweight, Py3.7+ dataflow engine for computational graph construction, manipulation, and distributed execution.
More information in this github issue

Snakebids: Snakemake + BIDS
Time slot : Atlantis
PROJECT DESCRIPTION
Snakemake (url, paper) is a fast-growing open source workflow management system that provides a high level of transparency, reproducibility and adaptability. Despite its wide adoption in the scientific community (>5 new citations per week), Snakemake has not yet had significant uptake in the neuroimaging community.
We have been working on an extension, Snakebids, to ease working with neuroimaging (specifically BIDS) data, and allow the creation of BIDS Apps with Snakemake (e.g. https://github.com/khanlab/hippunfold).
More information in this github issue
cleanBibImpact: Evaluating the impact of citation diversity statements
Time slot : Atlantis
PROJECT DESCRIPTION
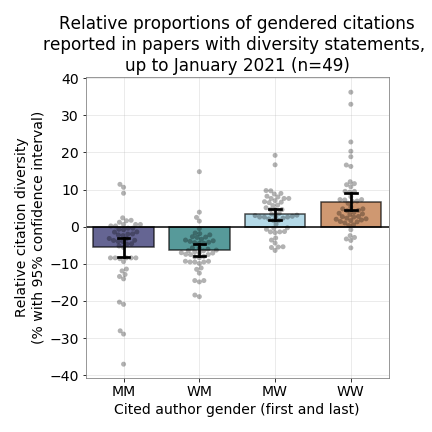
Gender imbalance is a big problem in academic research, and it is even reflected in our citation behavior. Our goal is to assess the 'impact' of the cleanBib tool, which was created to make researchers more aware of this problem. his tool can be used to assess the gender balance of authors in a reference list. Specifically, we want to find papers that cite the cleanBib code/paper/preprint, and see whether those citing papers have better gender balance n their reference lists than papers that do not.
In previous hackathons, we created this figure by manually collecting citation diversity data from papers with citation diversity statements. It shows the relative gender proportions in reference lists of papers with diversity statements (relative to the expected gender proportions).
More information in this github issue

Opscientia: Open Science Data Wallet
Time slot : Atlantis+Rising sun
PROJECT DESCRIPTION
The DataWallet is a dashboard connecting users to the decentralized web, and providing the option to validate and upload BIDS-compliant neuroimaging data, along with relevant metadata (Authors, paper, description, etc.) to peer to peer storage (IPFS). Data provenance and versioning is done using datalad with a special remote for IPFS.
More information in this github issue

Opscientia: Decentralised Open Science Stack
Time slot : Atlantis
PROJECT DESCRIPTION
Opscientia is a decentralised autonomous organisation (DAO) aiming to catalyse the pace of scientific discovery — starting with Neuroscience. Today, progress is limited by centralised systems built on outdated infrastructure. This restricts global access to study participation, research funding, scientific collaboration and knowledge creation. Participants also do not retain ownership or control over their own data. Our solution is a decentralised open science stack leveraging blockchain technologies. Global citizens are empowered to take part in studies whilst retaining full ownership and control over their data, and scientists are incentivised to share data and collaborate with their peers. Our solution empowers both citizens and scientists to become the new shareholders of scientific progress.
More information in this github issue
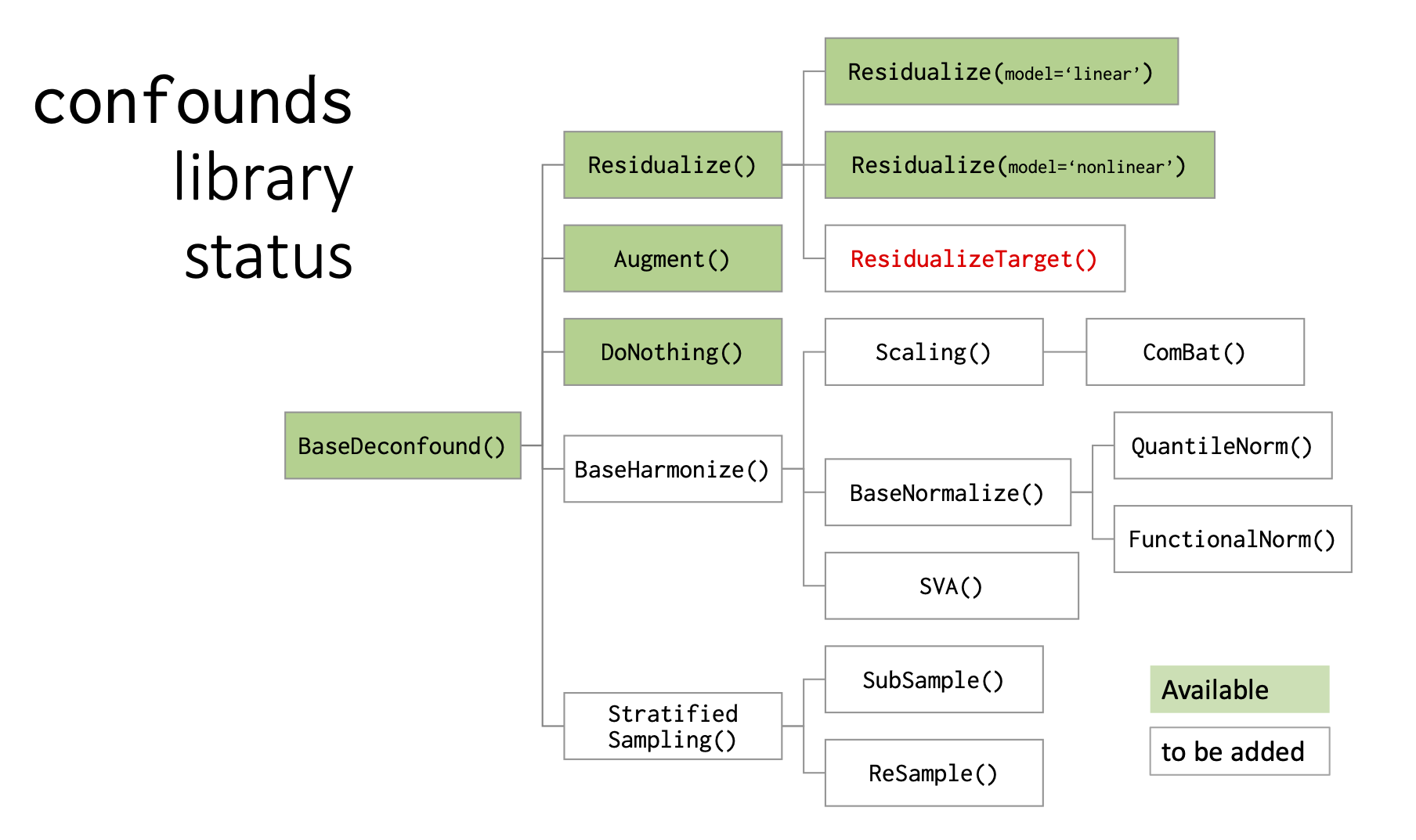
Python library to handle confounds/covariates: focus on ComBat
Time slot : Atlantis
PROJECT DESCRIPTION
Develop a python library of methods to handle confounds in various neuroscientific analyses, esp. statistics and predictive modeling. More info and slides here.
More information in this github issue

CONP: Sharing data and tools
Time slot : Atlantis
PROJECT DESCRIPTION
The Canadian Open Neuroscience Platform (CONP) provides an infrastructure for the promotion of open-science workflows and the sharing of neuroscience data. This platform brings together many of the country’s leading scientists in basic and clinical neuroscience to form an interactive network of collaborations in brain research, interdisciplinary student training, international partnership, clinical translation, and open publishing. The CONP aims to propel Canada’s basic and clinical neuroscience communities into a new era of commonly shared, digitally integrated, data- and algorithmic-rich neuroscience research.
More information in this github issue

Sea: A lightweight filesystem to process large neuroimaging data on HPC
Time slot : Atlantis
PROJECT DESCRIPTION
Large file transfers to and from slow storage can impede the processing of neuroimaging workflows processing such data. Sea is a file system that aims to reduce the costs of such file transfers by leveraging compute-local storage whenever possible during pipeline execution to offset the overhead of reading and writing to local storage.
More information in this github issue
About the Open-Science Special Interest Group
The OHBM Open Science SIG mission is to advance neuroimaging research by fostering the open sharing of ideas, data, and tools between members of the OHBM community. This will be accomplished by organizing educational events, supporting collaborative initiatives, and providing mentorship to junior researchers.As with previous years, our three main contributions to the OHBM annual meetings are:
- organizing a Brainhack to precede the annual meeting
- introducing the tools of open science via interactive tutorials as part of the TrainTrack
- facilitating collaborative work on a wide range of open science projects
This year the event is happening virtually, and we look forward to this opportunity to reach out to a larger and more diverse audience and to introduce a number of innovations that will enhance virtual collaborations!